Making the supertree: methods and data
Supertrees work by combining smaller trees – the results of numerous smaller analyses based on detailed investigations into the anatomy of various subgroups – together. Many dinosaur workers prefer an informal approach to supertree construction, in which the author or authors pick just a handful of trees that they feel are good representations of the relationships of those species and stitch them together by hand. There are definite advantages to this approach. It is very quick, for example, and allows the author(s) to avoid studies that gave spurious results or that have been superseded by the same authors.
The informal approach has so far been used in several studies of dinosaur evolution including size change, dietary adaptations and estimates of gaps in the record, whereas the earlier formal supertree was used in just one evolutionary study. However, it can be argued that the informal approach is not truly scientific: there is no optimality criterion (and so no measure of the supertree’s success), no explicit criteria for what is a ‘good’ and what is a ‘bad’ study (and so no justification for what is included and excluded) and no clear method for resolving conflicts in relationships between trees (and so no repeatability in the way the trees were stitched together).
The alternative to the informal approach is the formal approach, where protocols covering each step are explicitly defined at the outset in such a way that a computer could conceivably be programmed to complete the task. In reality, however, much of the steps of formal supertree reconstruction require a significant amount of worker effort (this project took four years from initiation to publication and involved nine people).
The first step in any formal supertree approach is to hunt through the primary literature (technical scientific publications found in specialist journals and books) and track down every family tree of the group under consideration. Not all of these trees can be considered equal, but choosing which to keep and which to disregard requires an explicit criterion. Here we chose trees that were the result of an explicit, numerical cladistic analysis and required that the published account include a list of the morphological characters used and a matrix describing their distribution in the taxa considered. However, many characters and even whole matrices are repeatedly reused.
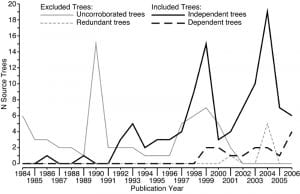 We also used the character lists to identify these and excluded trees that were redundant (i.e. had been replaced by a later study). In some cases multiple trees had equal claim to be the most recent version of a matrix and so we included them all, but weighted them in searches so their summed contribution was equal to one independent tree. In practice many of the trees used in the earlier supertree were excluded, but numerous more recent studies took their place. A graph of the trees used (and not used) by their year of publication is shown right:
We also used the character lists to identify these and excluded trees that were redundant (i.e. had been replaced by a later study). In some cases multiple trees had equal claim to be the most recent version of a matrix and so we included them all, but weighted them in searches so their summed contribution was equal to one independent tree. In practice many of the trees used in the earlier supertree were excluded, but numerous more recent studies took their place. A graph of the trees used (and not used) by their year of publication is shown right:
Making the supertree: taxonomy
The next big issue of supertree construction we had to tackle was taxonomy – the system of naming the dinosaurs. The problem here is that authors use different ranks of taxa in their trees (e.g. Tyrannosaurus rex – a genus and species, Tyrannosauridae – a family, Saurischia – an order). We can’t simply mix these trees together, for example Tyrannosaurus rex belongs inside both Tyrannosauridae and Saurischia. A protocol is required for replacing higher ranks (Saurischia, Tyrannosauridae) with their constituent members. Previous supertree analyses did this by simply using a taxonomic database, but here another novel approach was used that was inspired by Rod Page.
When we encountered a higher rank (e.g. Tyrannosauridae) we checked other trees where groups of species (clades) shared the same label and noted down which species were included and excluded in each case. Once this separate database was complete a list of appropriate replacements was available that relied not on taxonomy, but on the actual source trees – a much more appropriate approach.
Taxonomy also throws up another problem – not all dinosaur workers agree on how many species of dinosaur there are. Some species are named based on scrappy material that may or may not be diagnostic, whereas others are very similar to species that are already known and may actually belong in the same taxon. (For a review of these issues in dinosaurs see Benton 2008a, b). Teasing apart these issues is complex and controversial so we opted to use a major authoratative reference (the book The Dinosauria, second edition, 2004). This list of valid names was used to scour the source trees and remove or replace invalid names.
Ultimately 441 distinct valid species were included representing a sizeable chunk of known diversity (c. 70%). Only now were we ready to combine the trees.
Multiple supertree algorithms (computer routines) exist for linking trees together. These differ in the optimality criterion, that is, the code used by the computer to select the ‘best’ result, as well as the time they take to produce and particular oddities or biases in their results. Currently there is no clear favoured method so we chose to try several. Much of this work was done by Davide Pisani on a computer cluster (lots of powerful computers joined together) and a few supertrees were produced. Having pruned out some species which had too little information to place accurately (leaving 420) we then assessed each different supertree for how well it fitted the source trees.
The supertree, and our further macroevolutionary studies are developed further here.
References
Benton, M. J. 2008a. Fossil quality and naming dinosaurs. Biology Letters 4, 729-732. pdf.
Benton, M. J. 2008b. How to find a dinosaur, and the role of synonymy in biodiversity studies. Paleobiology 34, 516-533. pdf.

